Ongoing Projects
Ongoing Projects
Identification and validation of the neuronal substrates of Ube3a
Our research has focused on the identification of the neuronal substrates of Ube3a, the enzyme responsible for Angelman syndrome, using Drosophila melanogaster as a model organism (Ramirez et al., 2018). We aim to decipher the molecular mechanisms of Angelman syndrome combining innovative proteomic approaches with biochemistry, cellular and molecular biology in mice and human cells. We are currently working on the identification of the neuronal substrates ubiquitinated by Ube3a in mice, as well as in the validation of several candidate UBE3A substrates in human cell lines.
Characterization of the neuronal role conferred through ubiquitination by UBE3A
We have already confirmed the proteasome to be regulated by UBE3A in human cell culture (Ramirez et al., 2018), suggesting a mechanism for the complex symptoms of Angelman syndrome. We next want to elucidate the neurophysiological aspects of ubiquitination via UBE3A using primary neuronal cultures and acute slices as experimental model systems. We are aiming to understand the biological role of UBE3A in neurons, and so we will analyze the role of ubiquitinated substrates on critical indicators of excitability including integration and propagation of signals within the dendritic compartments of these neurons and their action potential output.
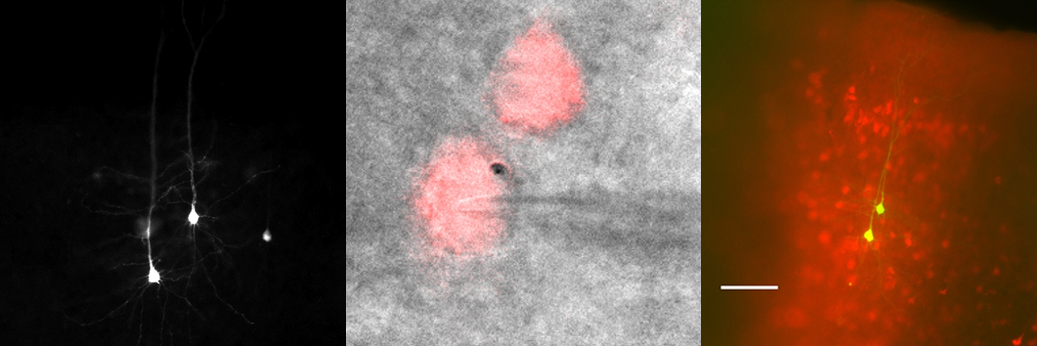
Investigating the effect of CaMKII on Angelman Syndrome
Mice models for Angelman syndrome show a deficit in long-term potentiation, a process in which Calmodulin-dependent kinase II (CaMKII) plays a crucial role. Interestingly, overexpression of CaMKII can rescue some synaptic aspects of Angelman Syndrome. While CaMKII is known to be ubiquitinated, UBE3A does not seem to be directly involved; so we now aim to determine the E3 ligase and the DUB enzyme regulating the ubiquitination of CaMKII. We also aim to determine, both in cells and flies, the proteins that interact with CaMKII, using proximity-dependent biotin identification.
Identification of DUB enzymes opposing UBE3A action
Deubiquitinating (DUB) enzymes exert the opposite effect to E3 ligases. We are trying to identify the DUB that specifically targets one of the substrates of UBE3A, and we want to determine whether inhibition of this DUB is sufficient to restore physiological ubiquitination levels of the UBE3A substrates. Additionally, we aim to test whether this inhibition could rescue the phenotype described for Angelman Syndrome in animal models, both in flies and mice.
Identification of UBE3A interactors and resolution of full-length UBE3A structure
The overall structure of UBE3A is still unknown. Human UBE3A was crystallized in 1999, but only its C terminal HECT domain structure was solved, together with its E2 partner, UbcH7. In order to decipher the molecular mechanisms of Angelman syndrome we are aiming to identify the neuronal interactors of UBE3A, using proximity-dependent biotin identification. Once we find those, we expect we might be able to solve the full-length crystal structure of UBE3A, which should then allow us to better understand the involvement of its N terminal part in its interaction with other proteins, including both cofactors and substrates.
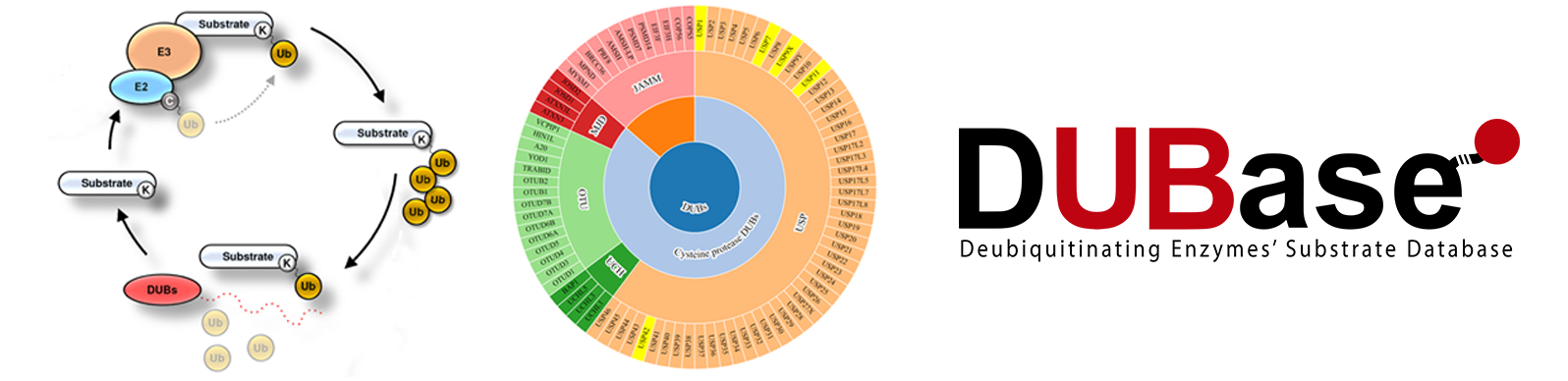
Global DUB substrate mapping
The post-translational modification of proteins with ubiquitin is a reversible process, with 80 human deubiquitinating enzymes (DUBs) being responsible for removing ubiquitin moieties from targeted proteins. Interest in the role of DUBs is rapidly growing, as they are the most likely component of the ubiquitin proteasome system pathway to be used as therapeutic targets. Within the context of the PRB2 Proteored, we are therefore aiming to generate a global map of DUB substrates in humans. Knowing the substrates regulated by each DUB will be a valuable resource for DUB-specific drug development in the future.
Identification of the neuronal substrates of other E3 ligases
We already demonstrated that the bioUb strategy can be applied to detect substrates of E3 ligases associated to neurological disorders such as Parkinson´s Disease (Martinez et al., 2017) or Angelman syndrome (Ramirez et al., 2018). Widening our collaborative efforts, we are currently carrying out the same approach to identify the substrates of other disease-linked E3 ligases.
Proteostasis deregulation in neurological rare diseases
Rare diseases have been generally understudied due to their low prevalence and their low value from a pharmaceutical market point of view. A significant number of those are known to be linked to misregulated proteostasis, so we are now aiming to identify the proteins whose physiological ubiquitination levels are altered in those rare neurological diseases. We expect that this research project will contribute towards a better understanding of the molecular causes underlying those diseases.