|
|
El formato de fichero y las APIs asociadas NetCDF/HDF5 se utiliza cada vez más en aplicaciones científicas dada su versatilidad y la posibilidad de utilizar técnicas de entrada/salida paralela. Además de ver las características de estos formatos de ficheros, se verá la forma de acceder desde C, FORTRAN y Python.
Datos del curso
- Donde: Santiago de Compostela.
- Inscripción: hasta el 7 de Septiembre de 2012.
- Plazas: 15.
- Coste: 60 €.
- Celebración: del 10 y 11 de Septiembre de 2012 en
Avda. de Vigo s/n, Campus Vida
Santiago de Compostela
Edficio CESGA
Más información
Página web del curso https://www.cesga.es/es/actuais/ver_curso/id_curso/2249
The Prof. Gernot Frenking, from the University of Marburg, will give a talk entitled Unusual Chemical Bonds and Reactivities – Connecting Fundamental Research with Application in Synthetic Chemistry.
Where: Donostia International Physics Center (DIPC), Donostia.
When: Next Friday, July 6th at 11.3o
Datos básicos
Cúando: del 2 al 12 de julio.
Donde: En la Universidad Politécnica de Valencia.
Horas lectivas: 32 horas, 5.5 horas de actividades no lectivas.
Cloud Computing
La computación en Nube (Cloud computing) permite externalizar los recursos de computación de una organización a un tercer proveedor, permitiendo un aprovisionamiento y liberación rápido de dichos recursos a través de Internet. Esta flexibilidad en la gestión de la computación, unida al modelo de pago por uso disponible en los Clouds públicos, supone un importante reto para las organizaciones, que deben conocer las ventajas de esta tecnología para decidir decantarse hacia su uso en aras de reducir costes de operación. De la misma forma, el almacenamiento de datos en la nube (Cloud storage) se orienta a ofrecer recursos de almacenamiento como servicio, implicando una forma de acceso y características específicas. Por otro lado, las tecnologías de Cloud Computing ayudan a realizar una gestión más eficiente de los centros de cálculo privados ya que permiten crear y gestionar nubes privadas. Estos Clouds tienen muchas de las características de los Clouds públicos generales, con la ventaja de que la información sensible permanece bajo el control de la organización.
Temas a desarrollar
Este curso trata las ventajas (e inconvenientes) del Cloud desde un punto de vista pragmático, enseñando las tecnologías subyacentes y estudiando lafuncionalidad ofrecida actualmente. Se realizan prácticas sobre Clouds privados y públicos para que el alumno conozca de primera mano la tecnología, utilizando en ambos casos las tecnologías más actuales. También se analizan casos de éxito de uso del Cloud tanto en entornos académicos como empresariales.
Más información
Información básica del curso, temas a desarrollar, matrícula e información de contacto en la página del curso
http://www.cfp.upv.es/formacion-permanente/cursos/cloud-computing–tecnologias-y-herramientas-para-trabajar-en-la-nube_idiomaes-menuupvtrue-cid30039.html
Introduction
Next Generation Sequencers produce a very large set of short DNA sequences that must be assembled to produce larger and useful sequences. ABySS and velvet are programs used to perform this assembly. In this article we compare both programs from the point of view of their performance, because for large data sets we have realized that they consume large amounts of computational resources.
Both programs are installed in the Computing Service of the University of the Basque Country (IZO-SGI) and this article summarizes the information and benchmarks that can be found in the ABySS and velvet web pages of the Service. The information shown here can be extended by reading the corresponding web pages of the Service. In the next sections we describe the environtment and write about the parallelization, execution time and the memory use, but the last one is the key one. Velvet is composed by two binaries that must be run to assemble the sequences: velveth and velvetg.
Nodes
The benchmarks shown in this post have been run in nodes with two E5645 Xeon processors (12 cores) at 2.4 GHz and memory up to 96 GB.
Parallelization
Both programs have been parallelized, i.e., the problem is divided in pieces that are solved by several processors at the same time, reducing in this way the computing time. Velvet is parallelized using the OpenMP (Open MultiProcessing) scheme while ABySS is parallelized with MPI (Message Passing Interface). This different approaches have different implications. OpenMP limits the parallelization to one node, while with MPI you can run the code in parallel in one or more nodes, so you can use more resources than with OpenMP.
Execution time
The execution time of both ABySS and velvet are small enough to run them without problems in professional clusters, therefore we are not going to focus on the particular results. We are going to mention that the efficiency of the velvet parallelization is quite poor. Velveth has a reasonable performance when running in 4 cores but not in a higher number of cores. Velvetg is even worse and, in fact, is better to run it in serial than in parallel. Nevertheless, ABySS parallelized well up to 8 cores, but for higher number of cores the efficiency drops down.
RAM memory use
The RAM memory is the limiting resource for ABySS and velvet because they need a lot. In the figure bellow we observe the evolution of the RAM used as a function of the number of the sequences. Notice the logarithmic scales. The measurements (their logarithms) have been fitted to straight lines.
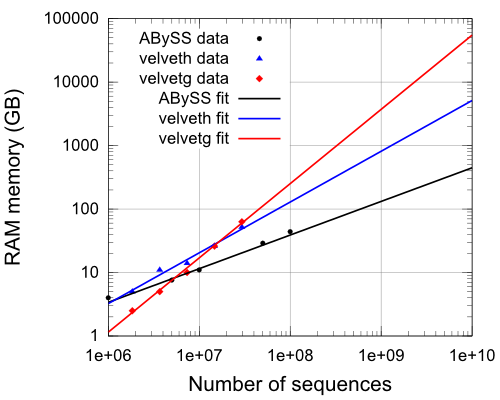
The equations of the fittings in the figure are:
R_velveth=(s^0.80)/19500
R_velvetg=(s^1.17)/(10^7)
R_abyss=(s^0.53)/450
Clearly, ABySS is better than velvet, as it uses much less memory. Velvet has smaller starting values but the memory grows almost linearly with the number of sequences. On the other hand, ABySS only grows more or less with the square root of the number of sequences and above 10^7 sequences it is more competitive than velvet in RAM terms.
This is specially important for large data samples. An assembly with 10^9 sequences can be done in a good cluster or server with ABySS, while is is nearly impossible to do it with velvet. Few computers in the world can afford to assign 4 TB of RAM to a single core in normal conditions.
In addition, due to the fact that ABySS is parallelized with MPI you can run ABySS in several nodes agregating their memory. If a lot of nodes are used to aggregate their RAM memory the execution time will be penalized due to the inefficiency of the parallelization for large number of cores, but probably this time increase can be affordable.
The second “HPC SysAdmin meeting” from HPCKP. This is a meeting aimed to share expertise and strategies in High Performance Computing & Clustering.
This meeting will be held at the University of Barcelona, from 15th October to 16th October, 2012. It is organized by the University of Barcelona (UB) and the Reference Network on Theoretical and Computational Chemistry (XRQTC).
Important Deadlines
| Abstract submission for presentations (tutorial or contributed talk) |
22th Sep 2012 |
| Confirm accepted presentations |
24th Sep 2012 |
| Early registration deadline for reduced fee |
30th Sep 2012 |
| Registration deadline |
07th Oct 2012 |
| Presentations submission deadline (PDF or PPT) |
10th Oct 2012 |
More information
Meeting web page: http://www.hpckp.org/index.php/anual-meeting/hpckp12
El Servicio General de Informática Aplicada a la Investigación (Cálculo Científico) ha abierto una convocatoria para seleccionar un nuevo técnico de apoyo con perfil en bioinformática, que será la candidata o candidato que se presente en la solicitud de ayuda para la contratación de personal técnico de apoyo ante el Ministerio de Economía y Competitividad.
Plazo de presentación de solicitudes del 11 al 15 de junio de 2012, ambos inclusive.
La convocatoria y bases así como otros detalles se encuentran en la página web de la UPV/EHU
http://www.pas-pertsonal.ehu.es/p263-content/es/contenidos/premio_concurso/12_pta_mec/es_pta_mec/pta_mec.html
y la correspondiente convocatoria es
PTA 06/12 – Informática Aplicada a la Investigación
Introducción
El CESGA en colaboración con la UDC convoca un curso de introducción al OpenACC. Pon tus viejos códigos C o FORTRAN a trabajar en entornos many-core o GPUs. Los computadores basados en unidades de procesamiento gráfico (GPUs) ofrecen una enorme potencia de cálculo con un consumo de potencia reducido. CUDA y OpenCL son los lenguajes más extendidos para la programación de GPUs. En los últimos años han surgido nuevos modelos de programación de GPUs basados en directivas de compilación. Las principales ventajas de estos modelos son las siguientes:
- Minimizar la reestructuración del código de la aplicación.
- Desarrollar aplicaciones para GPU independientes del hardware.
- Asegurar la portabilidad de las aplicaciones para GPU con las nuevas generaciones de hardware.
OpenACC:
- Es una iniciativa de estandarización para la programación de entornos many-core como las GPUs.
- Programación basada en directivas (como OpenMP), con lo que permite reaprovechar rápidamente el código existente en otros entornos de trabajo.
- Permite crear una única aplicación FORTRAN o C y ejecutarlas con o sin aceleradores en función de tus necesidades.
- Está apoyado por CAPS, NVIDIA, CRAY y PGI, lo que garantiza la portabilidad de tu aplicación a las futuras generaciones de GPUs.
- Permite disfrutar del rendimiento de las GPUs, sin necesidad de aprender lenguajes específicos ligados al fabricante, reduciendo el tiempo necesario para obtener resultados tangibles en tus proyectos de investigación o técnicos.
Datos de interés
Fechas del curso: del 25 al 27 de Junio
Horario del curso: de 10:00 a 14:00
Impartido por: Manuel C. Arenaz Silva
Coste matricula: 200€ IVA incluido.
Lugar de celebración: Avda. de Vigo s/n, Campus Vida – Santiago de Compostela – Edficio CESGA
Mas información y matrícula
Página web del curso https://www.cesga.es/actuais/ver_curso/id_curso/2242
OpenACC.
Introduction
Barcelona Computing Week 2012, July 2-6, at BSC/UPC, Barcelona.
Following the success of previous years, the Programming and tUning Massively Parallel Systems Summer School (PUMPS) offers researchers and graduate students a unique opportunity to improve their skills with cutting-edge techniques and hands-on experience in developing applications for many-core processors with massively parallel computing resources like GPU accelerators.
Participants will have access to a multi-node cluster of GPUs, and will learn to program and optimize applications in languages such as CUDA and OmpSs. Teaching assistants will be available to help with assignments.
Important information
- Applications due: May 31
- Notification of acceptance: June 15
- Summer school dates: July 2-6, 2012
- Location: Barcelona Supercomputing Center / Computer Architecture Dept. at Universitat Politecnica de Catalunya, Barcelona, Spain
Lecturers
- Wen-mei Hwu, University of Illinois at Urbana-Champaign
- David Kirk, NVIDIA Fellow, former Chief Scientist, NVIDIA Corporation
- Isaac Gelado, Rosa Badia, Xavier Martorell, Jesus Labarta, Nacho Navarro (BSC and UPC)
- Manuel Ujaldon (Univ. Malaga, CUDA Fellow in 2012)
The list of topics
- CUDA Parallel Execution Model
- CUDA Performance Considerations
- CUDA Algorithmic Optimization Strategies
- Data Locality Issues
- Dealing with Sparse and Dynamic data
- Efficiency in Large Data Traversal
- Reducing Output Interference
- Debugging and Profiling CUDA Code
- GMAC Runtime
- Multi-GPU Execution
- Introduction to OmpSs
- OmpSs: Leveraging GPU/CUDA Programming
- Hands-on Labs: CUDA Optimizations and OmpSs Programming
More information
Spring 2012 web seminars begin May 22nd and run through May 29th. As always, there is no cost to register for the talks, and the speakers will take questions from attendees. You can join the webcast seminars using both Windows and Mac computers. This spring talks will cover a variety of subject matter, including:
- BioLuminate, a protein-centric package for modeling biologics
- Advances in loop modeling and docking accuracy using GPCR structures and homology models
- What’s new in Schrödinger Suite 2012
- Popular additions to Canvas for advancing cheminformatics
More information
For registration and detailled information visit Schrödinger seminar center.
Details
The Schrödinger Workshops to take place on May 15th and 16th 2012 at CESCA facilities in Barcelona:
Centre de Serveis Científics i Académics de Catalunya, Gran Capitán , Annexus Building, 08034 Barcelona, Spain.
http://www.cesca.cat/
Topics
The first afternoon session is primarily an introduction and suitable to attend if you are a beginner, or have never worked with Schrödinger’s suite of programs.
The second day is a full day session for regular software users and beginners who attended the first session.
These workshops include theory and hands-on sessions to improve everyone’s skills on-site together with Jas Bhachoo, Senior Applications Scientist at Schrödinger.
The focus will be on structure-based and some ligand-based methods and their application in drug discovery.
Agenda
The agenda is as follows:
Tuesday the 15th of May 2012 – Afternoon Session 14.00-16.30
Learn How to Use Maestro
Using Maestro for Small Molecule Studies
How to:
- build a small molecule
- clean up a molecule with simple minimization
- run a conformational search
- use the Project Table for organizing your work
- superimpose two or more molecules (and to use the import panel)
- superimpose in the context of the protein environment
Using Maestro for A Simple Docking Study – Useful Workflows
- Protein Preparation
- Glide Grid Generation
- Glide Docking
- Post Docking Analysis with SiFTS
Wednesday the 16th of May 2012 – All day session 09.30-17.00
09.30 Welcome and Introduction
09.45 Session I:
Target structure prediction, refinement and characterization
Small molecule preparation with conformational searching
Virtual screening: Docking
13.00 Lunch Break
14.00 Session II:
Post docking analysis with clustering, SIFts, and extra-precision algorithms
Virtual screening: Structure-based pharmacophore screening and shaped-based screening
Data reduction and filtering using 2D approaches
16.30 Summary and Discussion
17.00 Close
Registration
This workshop is being offered free of charge but we ask you to register as space is limited. To register, please e-mail your name, title, institution to Katia Dekimeche Katia.Dekimeche@schrodinger.com and specify which day(s) you wish to attend.
|
|


Comentarios