Introduction
Next Generation Sequencers produce a very large set of short DNA sequences that must be assembled to produce larger and useful sequences. ABySS and velvet are programs used to perform this assembly. In this article we compare both programs from the point of view of their performance, because for large data sets we have realized that they consume large amounts of computational resources.
Both programs are installed in the Computing Service of the University of the Basque Country (IZO-SGI) and this article summarizes the information and benchmarks that can be found in the ABySS and velvet web pages of the Service. The information shown here can be extended by reading the corresponding web pages of the Service. In the next sections we describe the environtment and write about the parallelization, execution time and the memory use, but the last one is the key one. Velvet is composed by two binaries that must be run to assemble the sequences: velveth and velvetg.
Nodes
The benchmarks shown in this post have been run in nodes with two E5645 Xeon processors (12 cores) at 2.4 GHz and memory up to 96 GB.
Parallelization
Both programs have been parallelized, i.e., the problem is divided in pieces that are solved by several processors at the same time, reducing in this way the computing time. Velvet is parallelized using the OpenMP (Open MultiProcessing) scheme while ABySS is parallelized with MPI (Message Passing Interface). This different approaches have different implications. OpenMP limits the parallelization to one node, while with MPI you can run the code in parallel in one or more nodes, so you can use more resources than with OpenMP.
Execution time
The execution time of both ABySS and velvet are small enough to run them without problems in professional clusters, therefore we are not going to focus on the particular results. We are going to mention that the efficiency of the velvet parallelization is quite poor. Velveth has a reasonable performance when running in 4 cores but not in a higher number of cores. Velvetg is even worse and, in fact, is better to run it in serial than in parallel. Nevertheless, ABySS parallelized well up to 8 cores, but for higher number of cores the efficiency drops down.
RAM memory use
The RAM memory is the limiting resource for ABySS and velvet because they need a lot. In the figure bellow we observe the evolution of the RAM used as a function of the number of the sequences. Notice the logarithmic scales. The measurements (their logarithms) have been fitted to straight lines.
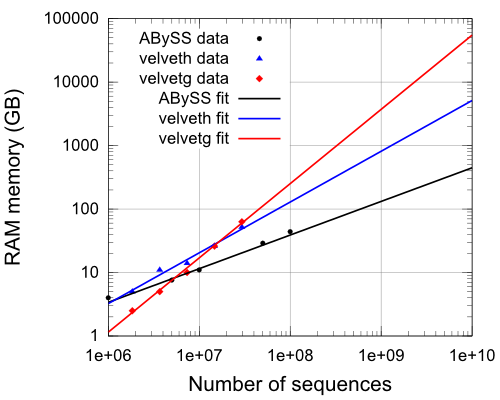
The equations of the fittings in the figure are:
R_velveth=(s^0.80)/19500
R_velvetg=(s^1.17)/(10^7)
R_abyss=(s^0.53)/450
Clearly, ABySS is better than velvet, as it uses much less memory. Velvet has smaller starting values but the memory grows almost linearly with the number of sequences. On the other hand, ABySS only grows more or less with the square root of the number of sequences and above 10^7 sequences it is more competitive than velvet in RAM terms.
This is specially important for large data samples. An assembly with 10^9 sequences can be done in a good cluster or server with ABySS, while is is nearly impossible to do it with velvet. Few computers in the world can afford to assign 4 TB of RAM to a single core in normal conditions.
In addition, due to the fact that ABySS is parallelized with MPI you can run ABySS in several nodes agregating their memory. If a lot of nodes are used to aggregate their RAM memory the execution time will be penalized due to the inefficiency of the parallelization for large number of cores, but probably this time increase can be affordable.

http://www.vouchercodeslug.co.uk
HPC @UPV/EHU » Benchmarking genetic assemblers: ABySS vs. velvet